Cardiotensor
( 
The library is designed for modern high-resolution modalities such as synchrotron tomography (HiP-CT), micro-CT, and 3D optical imaging. It supports datasets up to teravoxel scale through chunk-based and parallelized processing pipelines.
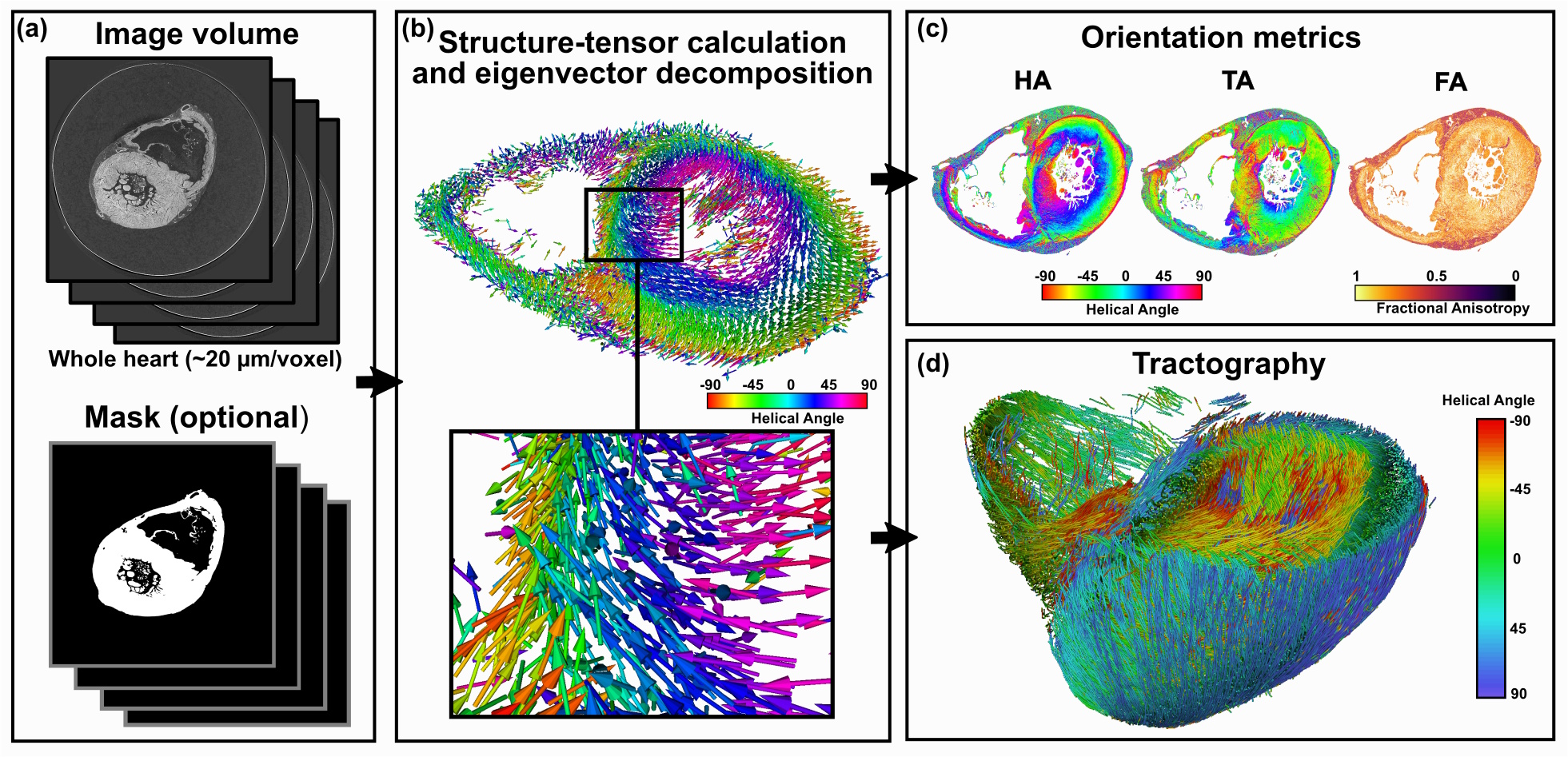
Cardiotensor workflow: orientation computation with structure tensor, helical angle (HA) and intrusion angle (IA) mapping, and streamline-based tractography
Key Features
- 3D Structure Tensor Analysis: Computes voxel-wise cardiomyocyte orientation from image intensity gradients.
- Cardiac Microstructure Metrics: Extracts helical angle (HA), intrusion angle (IA), and fractional anisotropy (FA).
- Tractography: Reconstructs cardiomyocyte trajectories for continuous myoaggregate mapping.
- Scalable Processing: Optimized for HPC clusters and terabyte-sized volumes using chunked parallelization.
- Visualization: Interactive 3D rendering of vector fields and streamlines, plus VTK export for ParaView.
- Tissue-Agnostic: Applicable beyond the heart to other fibrous tissues such as brain white matter, skeletal muscle, and tendon.
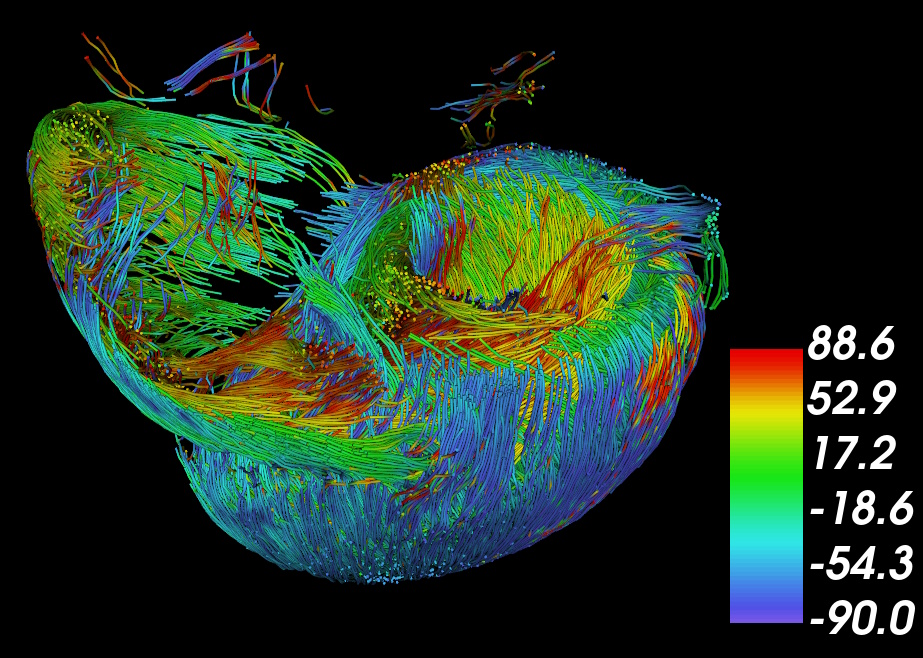
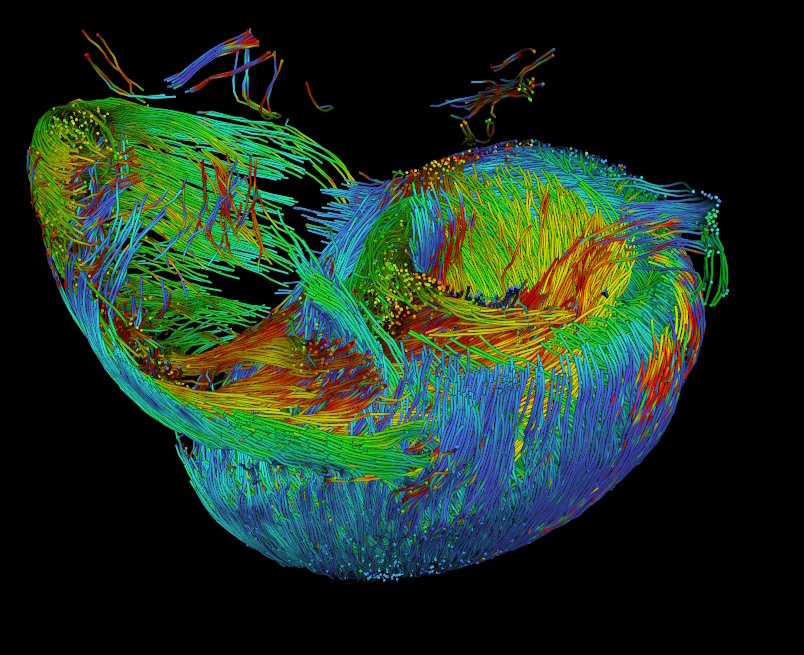
Streamline tractography of cardiomyocyte orientation, color-coded by helical angle (HA)
Why Cardiotensor?
Most established orientation-analysis tools (e.g., MRtrix3, DIPY, DSI Studio) were developed for diffusion MRI. Cardiotensor instead derives orientation directly from imaging intensity gradients, enabling analysis across modalities such as synchrotron tomography, micro-CT, and optical microscopy. Unlike small-scale in-house codes, it is designed to be scalable, reproducible, and openly accessible.
Documentation and Availability
📖 Full documentation, tutorials, and example datasets are available at:
https://josephbrunet.github.io/cardiotensor
Install via pip:
pip install cardiotensor
Acknowledgement
📄 For full details, see the preprint paper (2025).